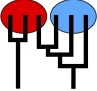
We finally published our divergence estimation method
Population divergence time estimation using individual lineage label switching., Download Migrate 4.4.x or 5.x and test for yourself.

Population divergence time estimation using individual lineage label switching., Download Migrate 4.4.x or 5.x and test for yourself.

Somayeh Mashayekhi (postdoc until 2019, now Assistant Professor at Kennesaw State University GA) developed a generalization of Kingman's coalescent.

Kyle Shaw extended currently work of Justin Bricker integrating sequencing errors with the Poisson-indel process.

Haleh Ashki developed a method to use the genetic information of the pathogen to select among candidate contact networks of the host.

Haleh Askhi developed simulation methods for dynamic networks in epidemiology that allows fast simulations of epidemics on complex, changing contact networks.